Community Tools for Analysis of NASA Earth Observation System Data in the Cloud

EOSDIS SE TIM 2019
Andrew Pawlosk andrew@element84.com
Talk Agenda
- Pangeo 101
- Jupyter + Xarray + Dask
- STAC + Intake + EOSDIS
- Lessons Learned
What is Pangeo?
- Not a software distro, package, or library
- Community of data users and software developers
- Frequently used packages (Pangeo "stack")
- Curated set of tools, architectures, and methodologies
 Image Source: Jake VanderPlas, "The State of the Stack," SciPy Keynote (SciPy 2017).
Image Source: Jake VanderPlas, "The State of the Stack," SciPy Keynote (SciPy 2017).
 Image Source: Theo McCaie, UK Met Office, ESA Φ-week Event.
Image Source: Theo McCaie, UK Met Office, ESA Φ-week Event.
Goal:
Foster collaboration around open source scientific python ecosystem for ocean / atmosphere / climate science
Goal:
Build out stack with domain specific packages (e.g. thermodynamics, regridding, vector calculus, etc)
Goal:
Scale to handle many-PB data missions (e.g. SWOT, NISAR)
EOSDIS Data Ingest Rates
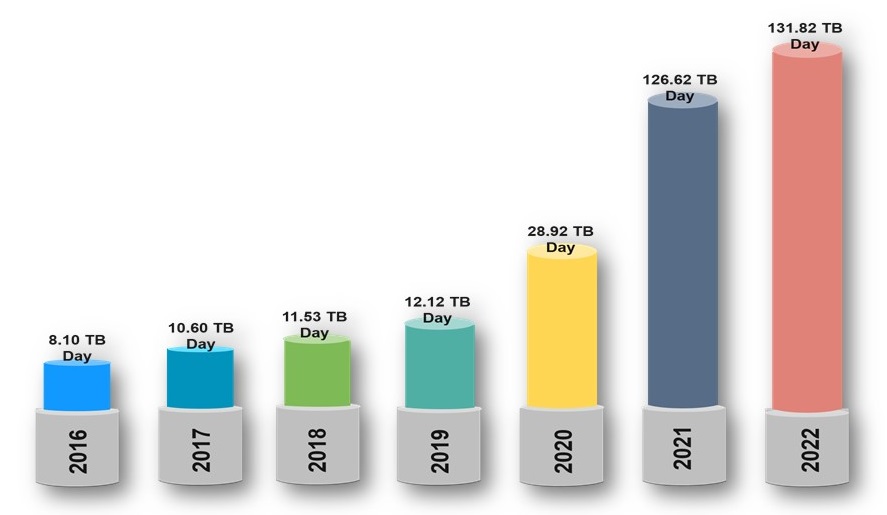 Image Source: https://earthdata.nasa.gov/about/cloud-evolution
Image Source: https://earthdata.nasa.gov/about/cloud-evolution
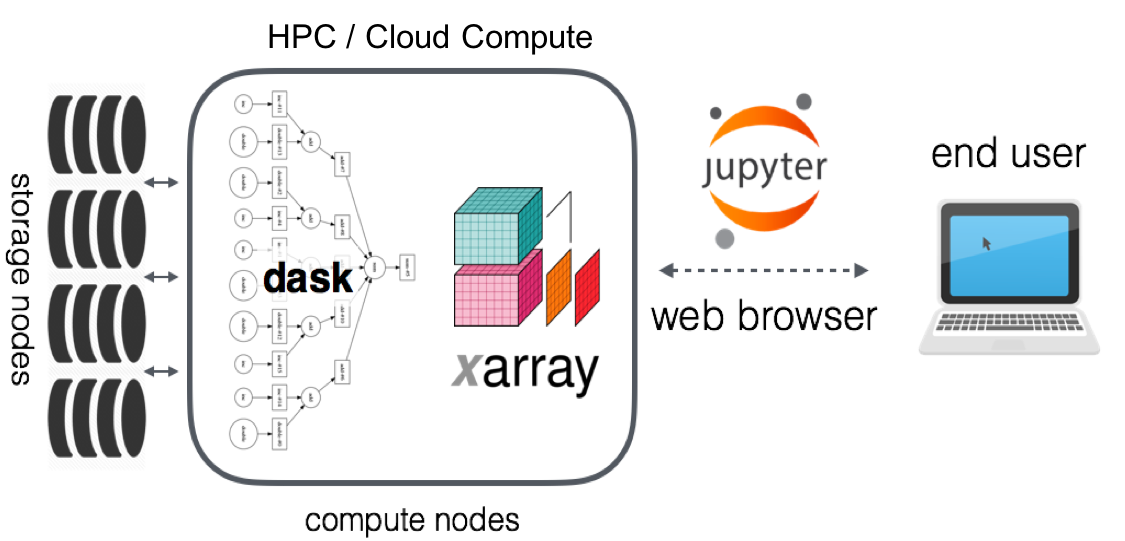 Image Source: Pangeo Technical Architecture (https://pangeo.io/architecture.html)
Image Source: Pangeo Technical Architecture (https://pangeo.io/architecture.html)
- No source data downloaded to workstation
- Well suited for cloud-native data stores/formats (e.g. S3, GCS and COGs, Zarr)
- Notebooks can be persisted, shared
- Compute is ephemeral
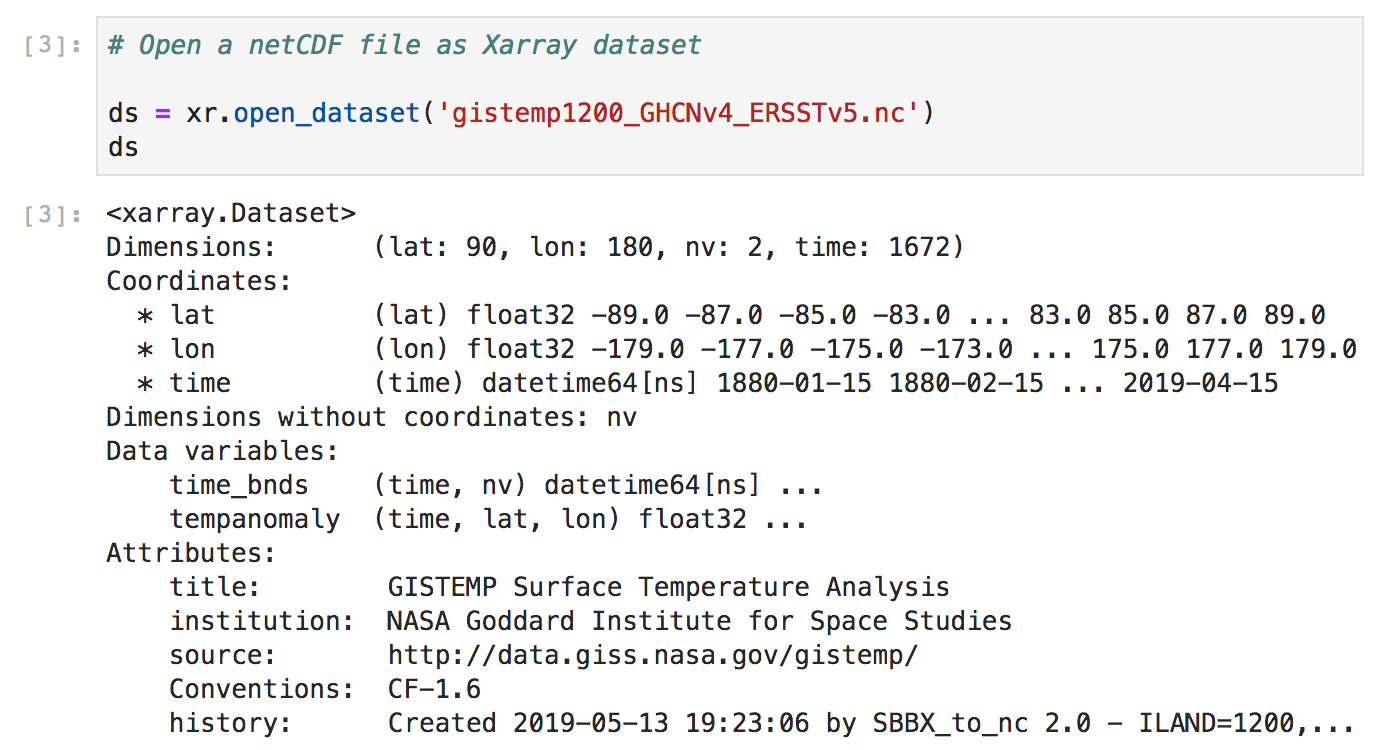
Xarray
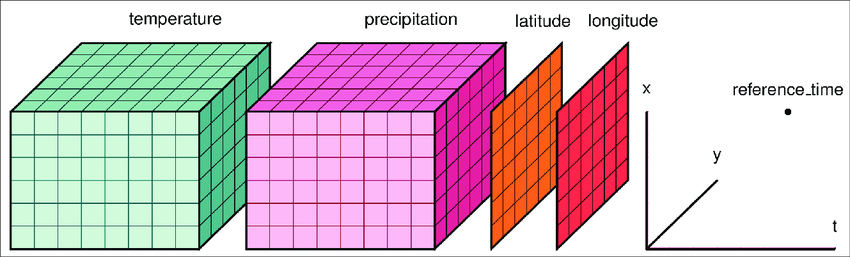 http://xarray.pydata.org/en/stable/api.html
http://xarray.pydata.org/en/stable/api.html
Xarray
Dask
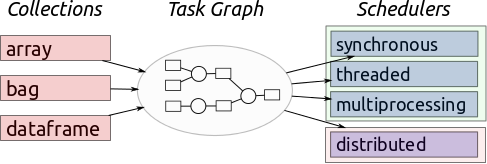 Image Source: Multidimensional Arrays, Geohackweek 2016
Image Source: Multidimensional Arrays, Geohackweek 2016
Dask
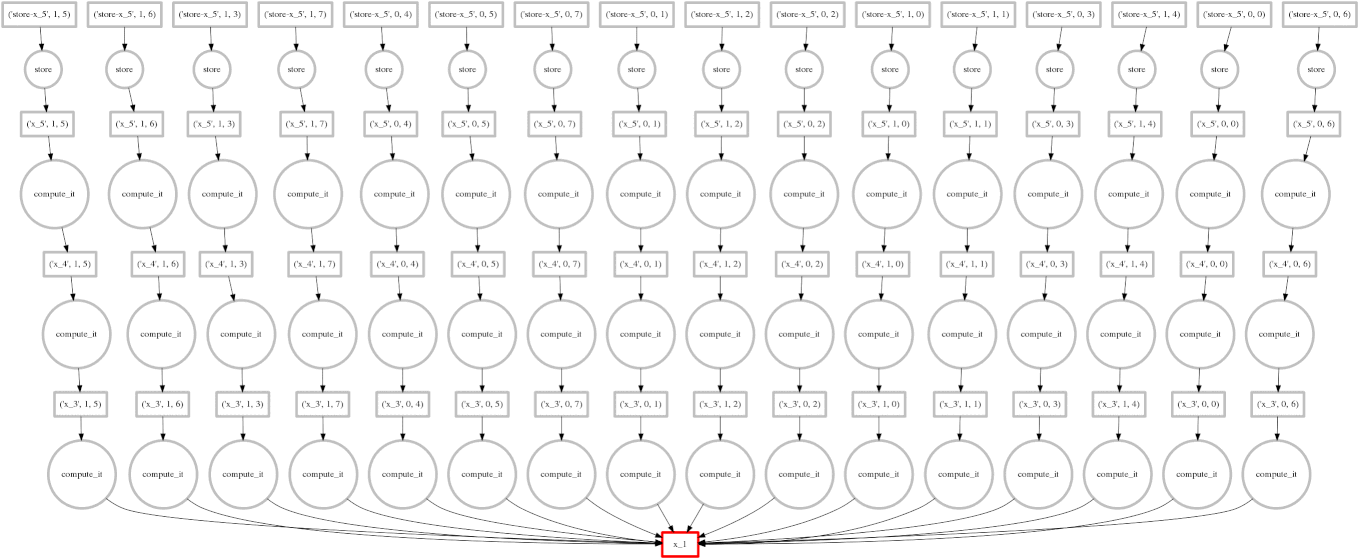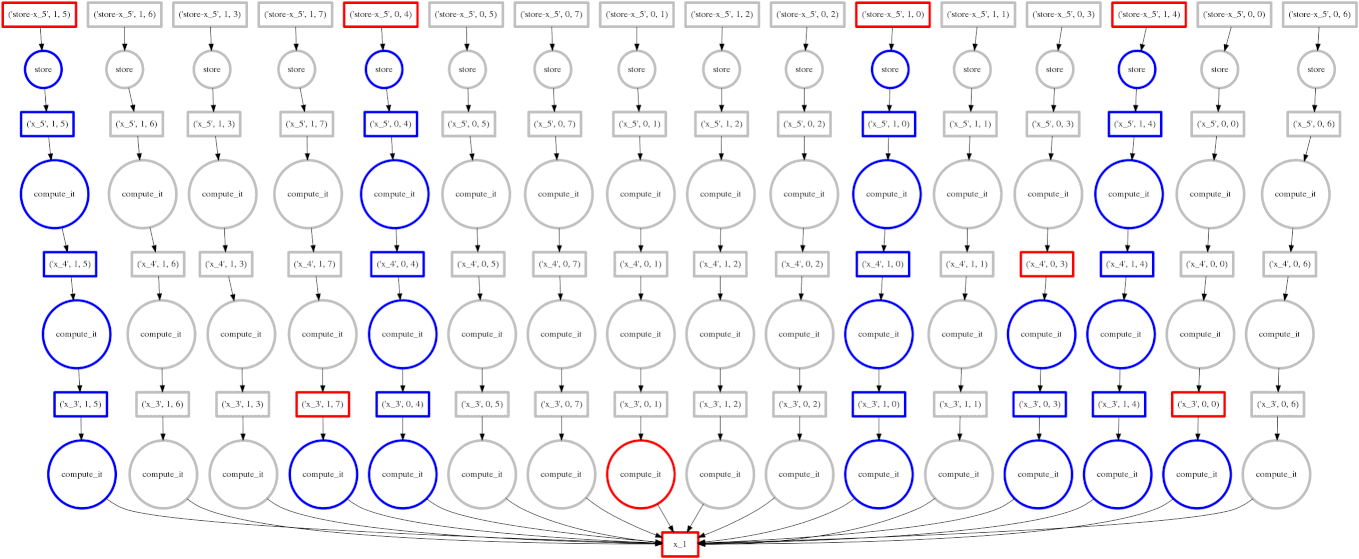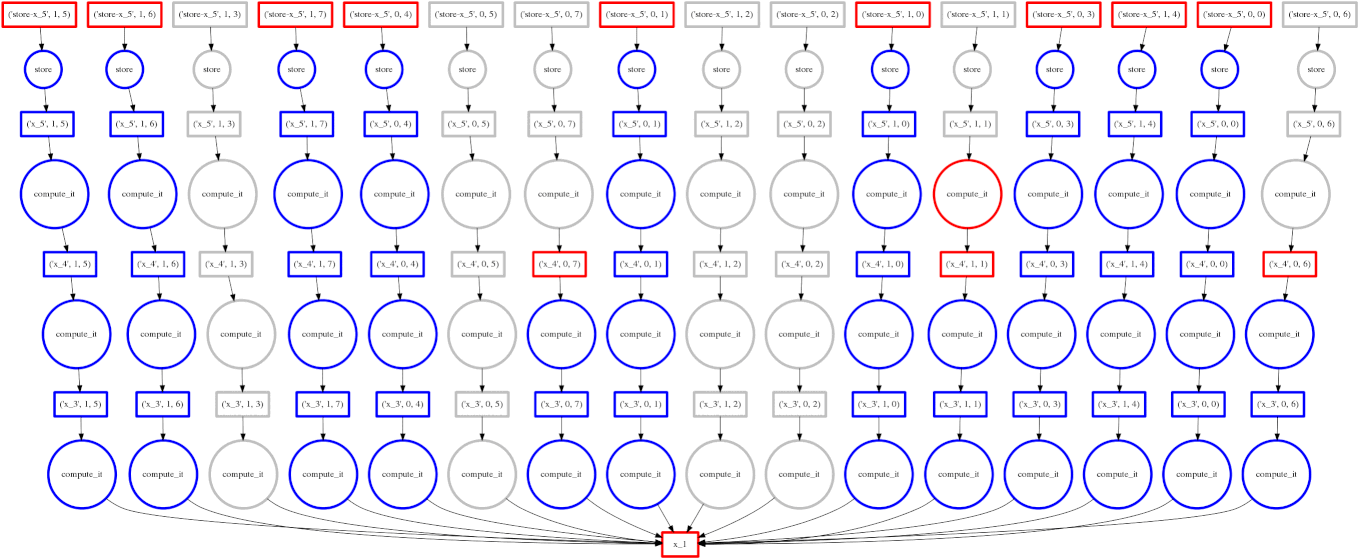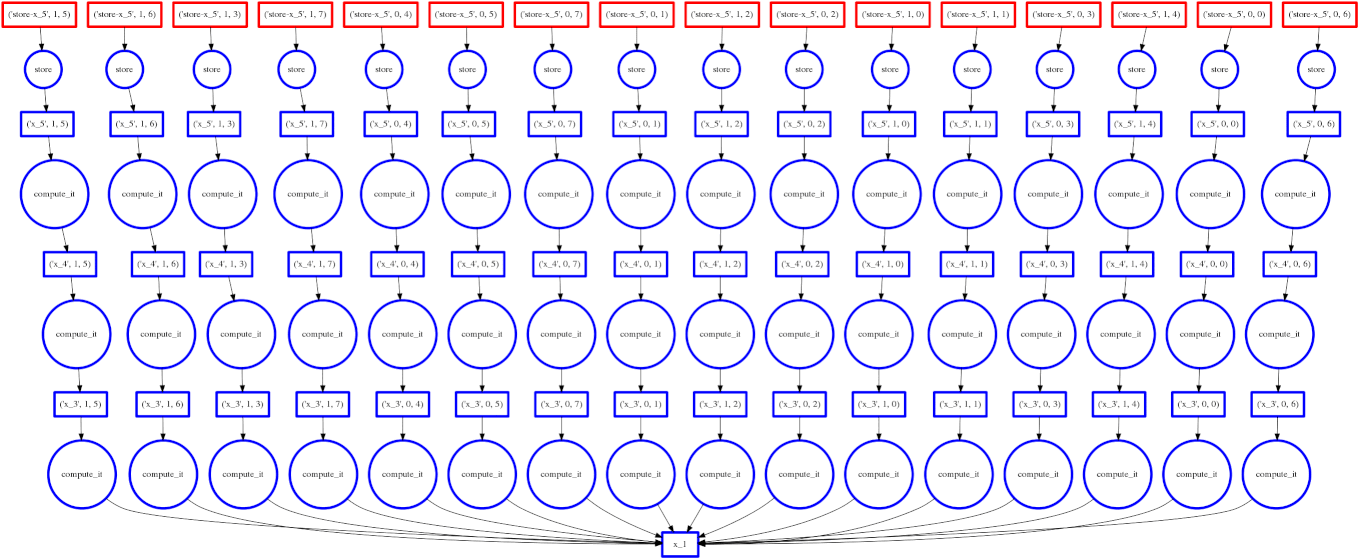 Image Source: Multidimensional Arrays, Geohackweek 2016
Image Source: Multidimensional Arrays, Geohackweek 2016
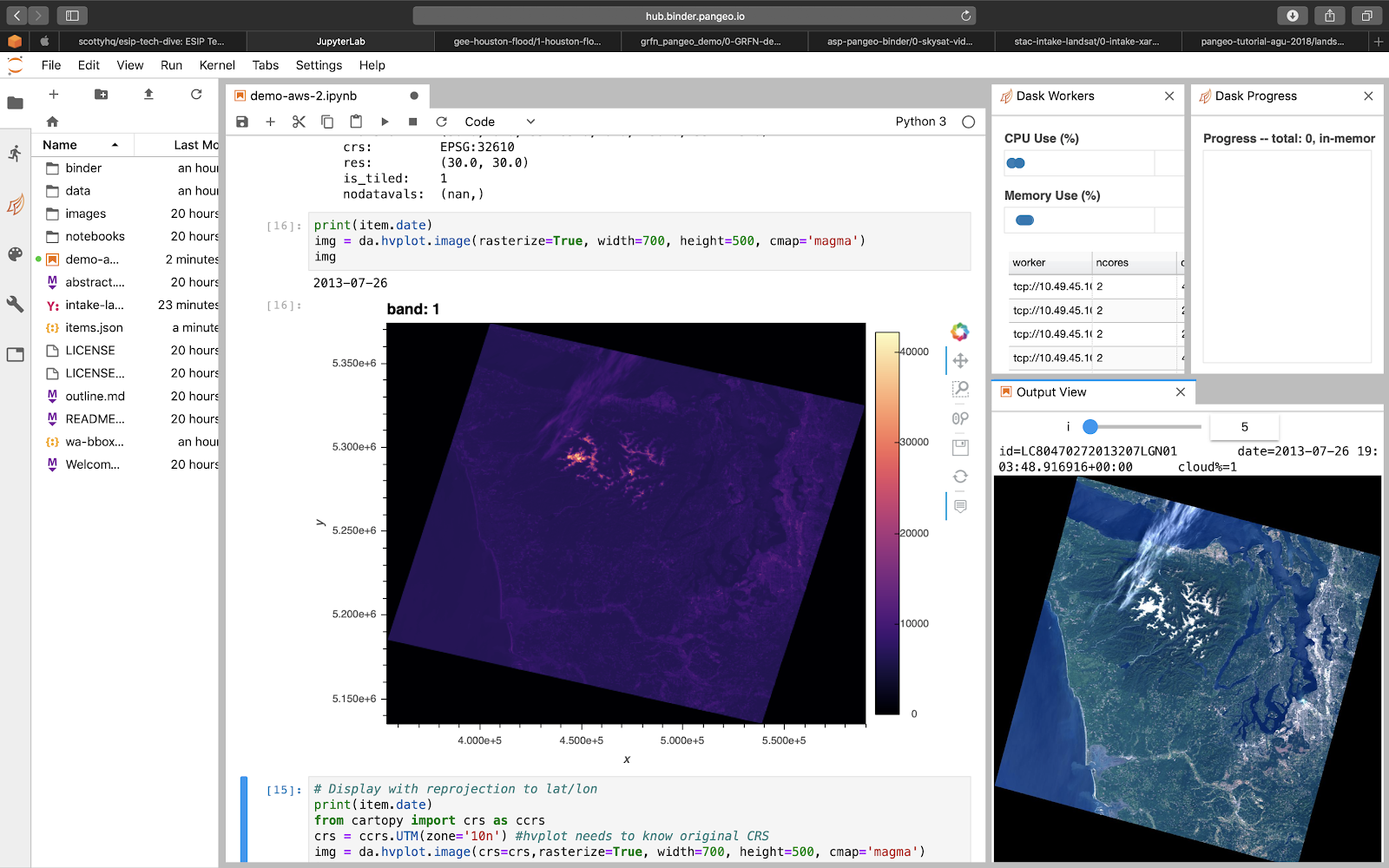 Image Source: Scott Henderson (University of Washington)
Image Source: Scott Henderson (University of Washington)
Jupyter is great for sharing code and coupling dependencies.
To be truly reproducible artifacts, notebooks must also share data.
Challenges remain for data access
- Notebooks often have hardcoded, brittle download mechanisms
- Data isn't always well-described
- Can involve repetitive boilerplate code


Intake
- https://github.com/intake/intake
- Abstracts data access mechanisms
- Opens data directly as python objects
- Extensible into different drivers
- Couple data like you would dependencies
STAC
- https://stacspec.org
- SpatioTemporal Asset Catalog
- Describes information about the Earth in a certain space and time
- Describes Items, Catalogs, and Collections
- Defines a RESTful API
- Highly extensible
Intake-Stac
- https://github.com/pangeo-data/intake-stac
- Imports STAC-described data directly into Jupyter kernel
- Opens as ready-to-use Xarray object(s)
# converting to intake catalog will enable intake tools such as gui browser
cat = intake.StacCatalog('landsat8-aws.json')
# or leverage existing tools such as sat-api/sat-search
cat = intake.StacSearch(collection='landsat8', bbox=[], datetime='2017/2019')
cat.filter(bands=['red','green','nir'], cloudcover=20)
# load as xarray dataset:
ds = cat.to_dask()
# need to share STAC catalogs with colleagues / reproduce work later
cat.to_file('my-catalog.json')
# converting to intake catalog will enable intake tools such as gui browser
cat = intake.StacCatalog('landsat8-aws.json')
# or leverage existing tools such as sat-api/sat-search
cat = intake.StacSearch(collection='landsat8', bbox=[], datetime='2017/2019')
cat.filter(bands=['red','green','nir'], cloudcover=20)
# load as xarray dataset:
ds = cat.to_dask()
# need to share STAC catalogs with colleagues / reproduce work later
cat.to_file('my-catalog.json')
# converting to intake catalog will enable intake tools such as gui browser
cat = intake.StacCatalog('landsat8-aws.json')
# or leverage existing tools such as sat-api/sat-search
cat = intake.StacSearch(collection='landsat8', bbox=[], datetime='2017/2019')
cat.filter(bands=['red','green','nir'], cloudcover=20)
# load as xarray dataset:
ds = cat.to_dask()
# need to share STAC catalogs with colleagues / reproduce work later
cat.to_file('my-catalog.json')
# converting to intake catalog will enable intake tools such as gui browser
cat = intake.StacCatalog('landsat8-aws.json')
# or leverage existing tools such as sat-api/sat-search
cat = intake.StacSearch(collection='landsat8', bbox=[], datetime='2017/2019')
cat.filter(bands=['red','green','nir'], cloudcover=20)
# load as xarray dataset:
ds = cat.to_dask()
# need to share STAC catalogs with colleagues / reproduce work later
cat.to_file('my-catalog.json')
Pangeo, Intake, STAC, and EOSDIS
- CMR will support STAC
- NSIDC STAC and cloud-native pilot efforts
EOSDIS data can be searched, accessed, and injected directly into Jupyter environments.
Science can be bundled, shared, and reproduced across the Pangeo ecosystem and beyond.
Thank you.


Further Resources
- Website: http://pangeo.io/
- Github: https://github.com/pangeo-data
- Blog: https://medium.com/pangeo
- Gitter: https://gitter.im/pangeo-data
Anthony Arendt (arendta@uw.edu)
Rob Fatland (rob5@uw.edu)
Joe Hamman (jhamman@ucar.edu)
Matt Hanson (mhanson@element84.com)
Scott Henderson (scottyh@uw.edu)
Dan Pilone (dan@element84.com)
Andrew Pawloski (andrew@element84.com)
Amanda Tan (amandach@uw.edu)
Backup Slides
Lessons Learned
Lessons Learned: Data Access
- Hitting S3 directly was by far the easiest way to operate
- Best performance when HTTP Range Header supported (e.g. Zarr, COGs)
Lessons Learned: Kubernetes on AWS
- EKS is great, but SDK wasn't the right choice out of the box
- Ultimately used eksctl for operations (autoscaling)
- Potential for fine-grained costs controls, but not there yet
Using Your Deployment: Dask
from dask_jobqueue import PBSCluster
from dask.distributed import Client
cluster = PBSCluster(cores=36,
memory="108GB")
cluster.scale(10)
client = Client(cluster)
Using Your Deployment: Dask
from dask_kubernetes import KubeCluster
from dask.distributed import Client
cluster = KubeCluster(n_workers=10)
cluster.scale(10)
client = Client(cluster)
Cloud-Native Data Formats
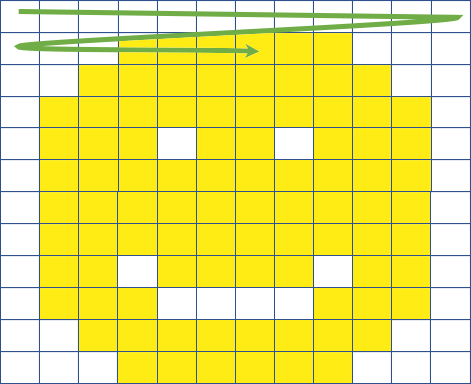
Image Source: James Norton (Element 84)
Cloud-Native Data Formats
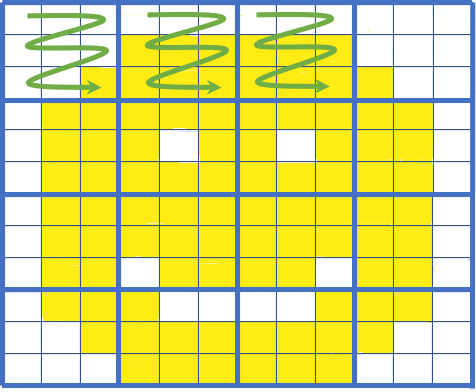
Image Source: James Norton (Element 84)

Cloud Optimized GeoTIFFs (COGs)
- Regular GeoTIFFs
- Tiled
- Support HTTP GET Range Requests
- End users download subset range of the GeoTIFF



Zarr
- Multi-dimensional arrays saved in discrete chunks
- Each chunk is a file
- Clients can pull only the chunks they need